Many years ago I started making videos of my lectures for my ecological statistics course. I still use those early videos, now on YouTube, but a recent comment on the checking assumptions video made me realize that I somehow misplaced the code that I used to create the figures in the powerpoints of the early videos. 1
Rather than try to reproduce the old code in a non-public way, I thought I would redo it here so it is easy to share. Plus, I get to show of the new function my students and I wrote for making nice residual plots with ggplot2. The exercise uses the horseshoe crab satellite male data from Agresti’s book on categorical data analysis. If you would like the explanations of the figures have a look at the video.
# install.packages("glmbb") # install Ben Bolker's package
data(crabs, package = "glmbb")
library("tidyverse")
library("broom")
library("NRES803") # devtools::install_github("atyre2/NRES803")The video doesn’t show it (it’s in another video), but we first need to fit a linear model of female weight in grams as a function of carapace width in cm.
hsc1 <- lm(weight~width, data = crabs)
summary(hsc1)##
## Call:
## lm(formula = weight ~ width, data = crabs)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1186.00 -116.15 -5.13 150.76 1015.50
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -3944.019 255.027 -15.46 <2e-16 ***
## width 242.642 9.666 25.10 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 267.4 on 171 degrees of freedom
## Multiple R-squared: 0.7865, Adjusted R-squared: 0.7853
## F-statistic: 630.1 on 1 and 171 DF, p-value: < 2.2e-16The first figure in the video is a histogram of the residuals, with the normal distribution superimposed.
hist(residuals(hsc1), main = "Histogram of Residuals",
freq = FALSE, col = NULL,
xlab = "Residuals")
# add the normal curve with the residual standard error from model
curve(dnorm(x, sd = 264.1), from = -1000, to =1000, add = TRUE)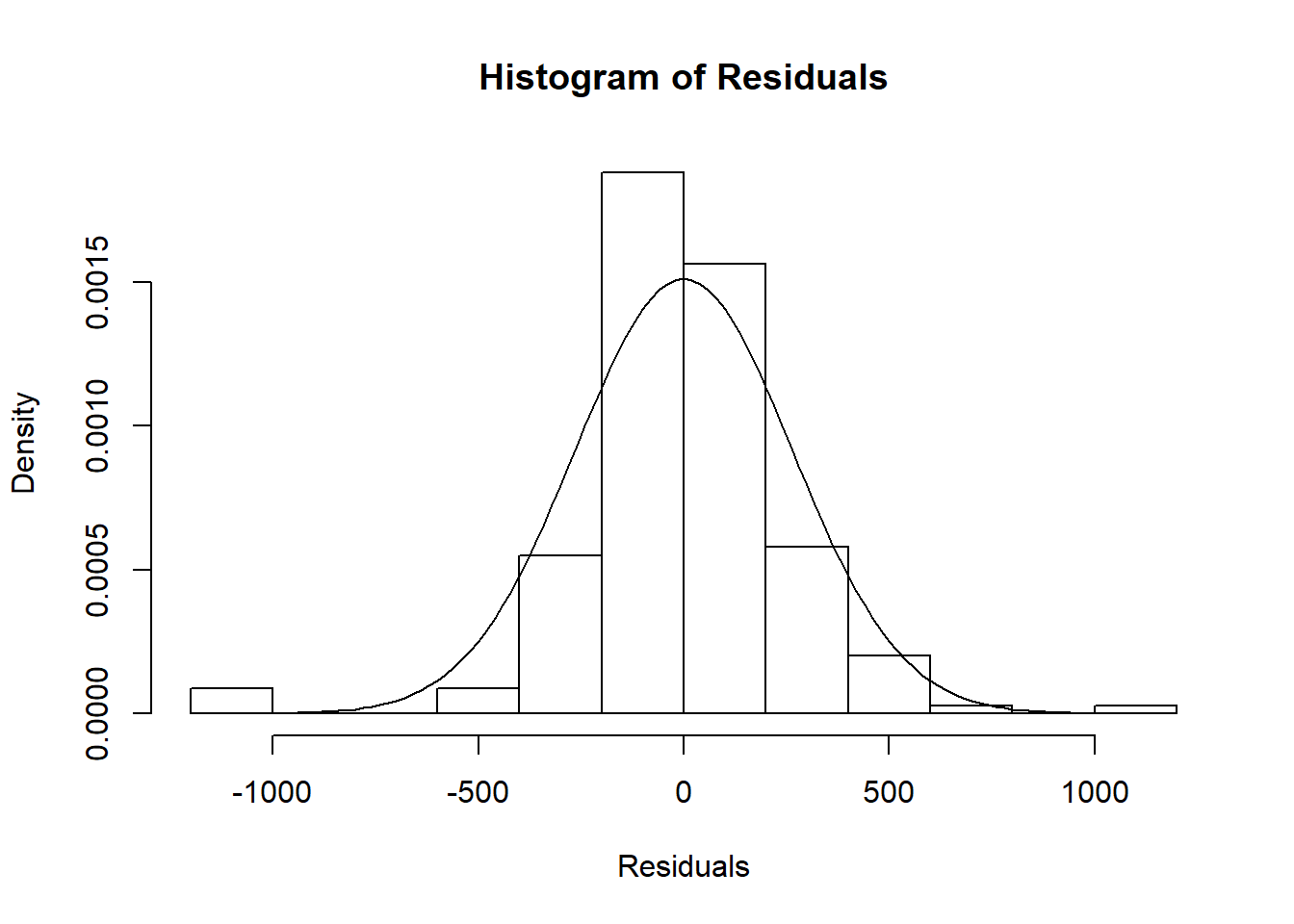
In the video I also mention the results of a Kolmogorov-Smirnov test of the residuals vs. a normal distribution.
ks.test(residuals(hsc1), "pnorm", 0, 264.1)## Warning in ks.test(residuals(hsc1), "pnorm", 0, 264.1): ties should not be
## present for the Kolmogorov-Smirnov test##
## One-sample Kolmogorov-Smirnov test
##
## data: residuals(hsc1)
## D = 0.089237, p-value = 0.1271
## alternative hypothesis: two-sidedThen there’s a quantile-quantile plot.
qqnorm(residuals(hsc1))
qqline(residuals(hsc1))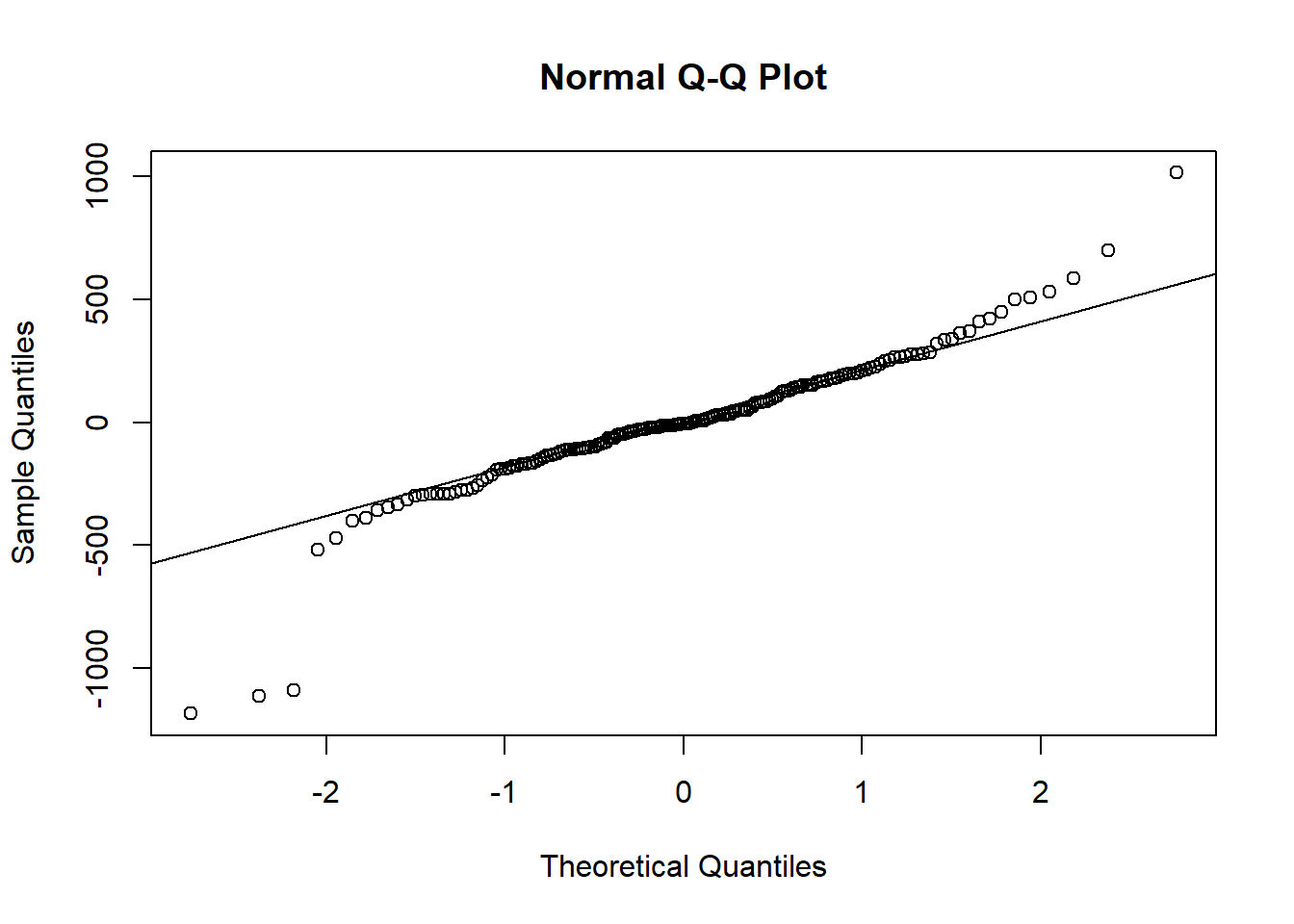
Next is the output of the plot() on a linear model object.
old.par <- par(mfrow = c(2,2))
plot(hsc1)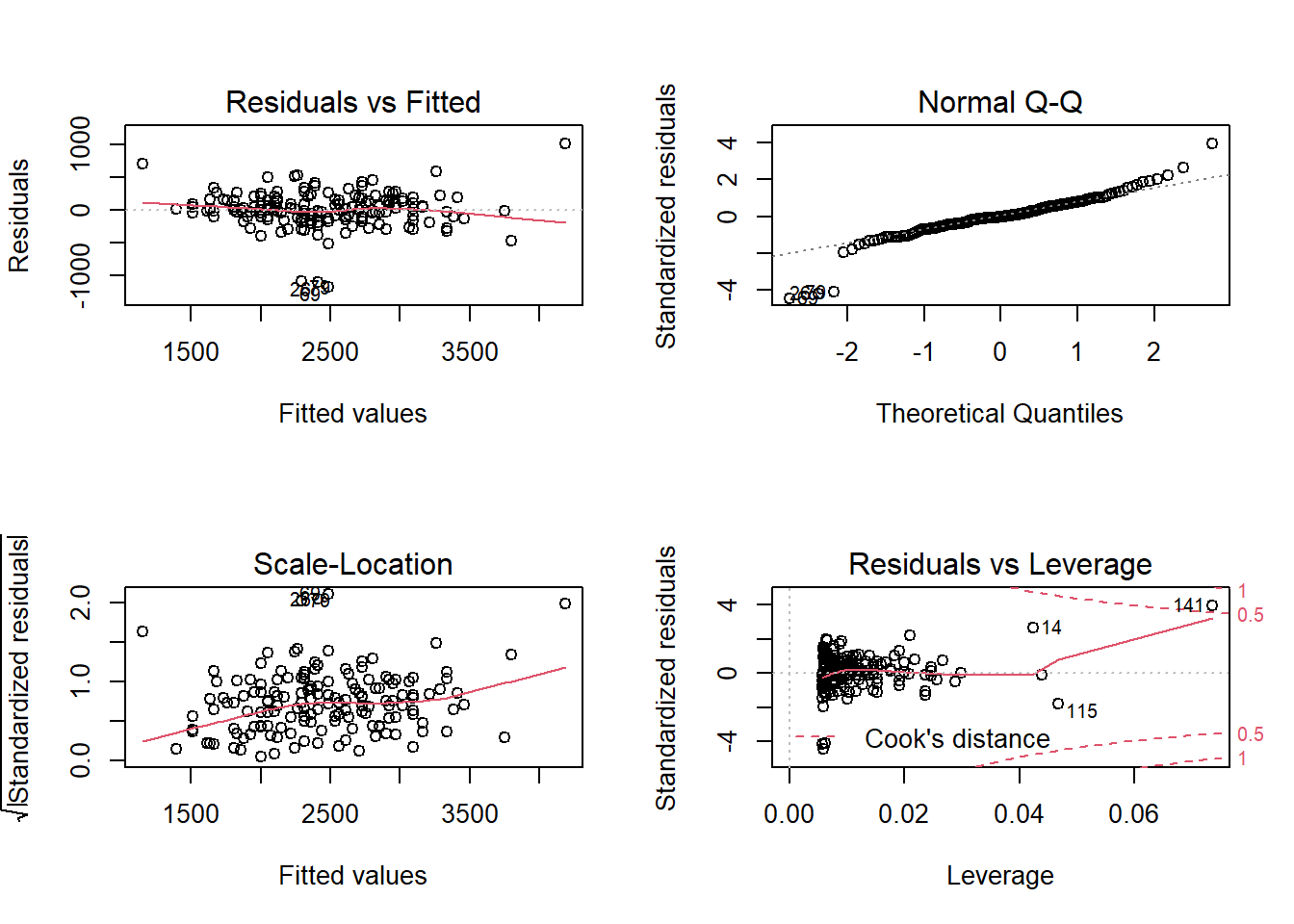
par(old.par)My students and I put together a function that uses ggplot2 to make something similar. I like it because it includes confidence regions, which makes it easier to judge if the smooth spline is “flat”.
check_assumptions(hsc1)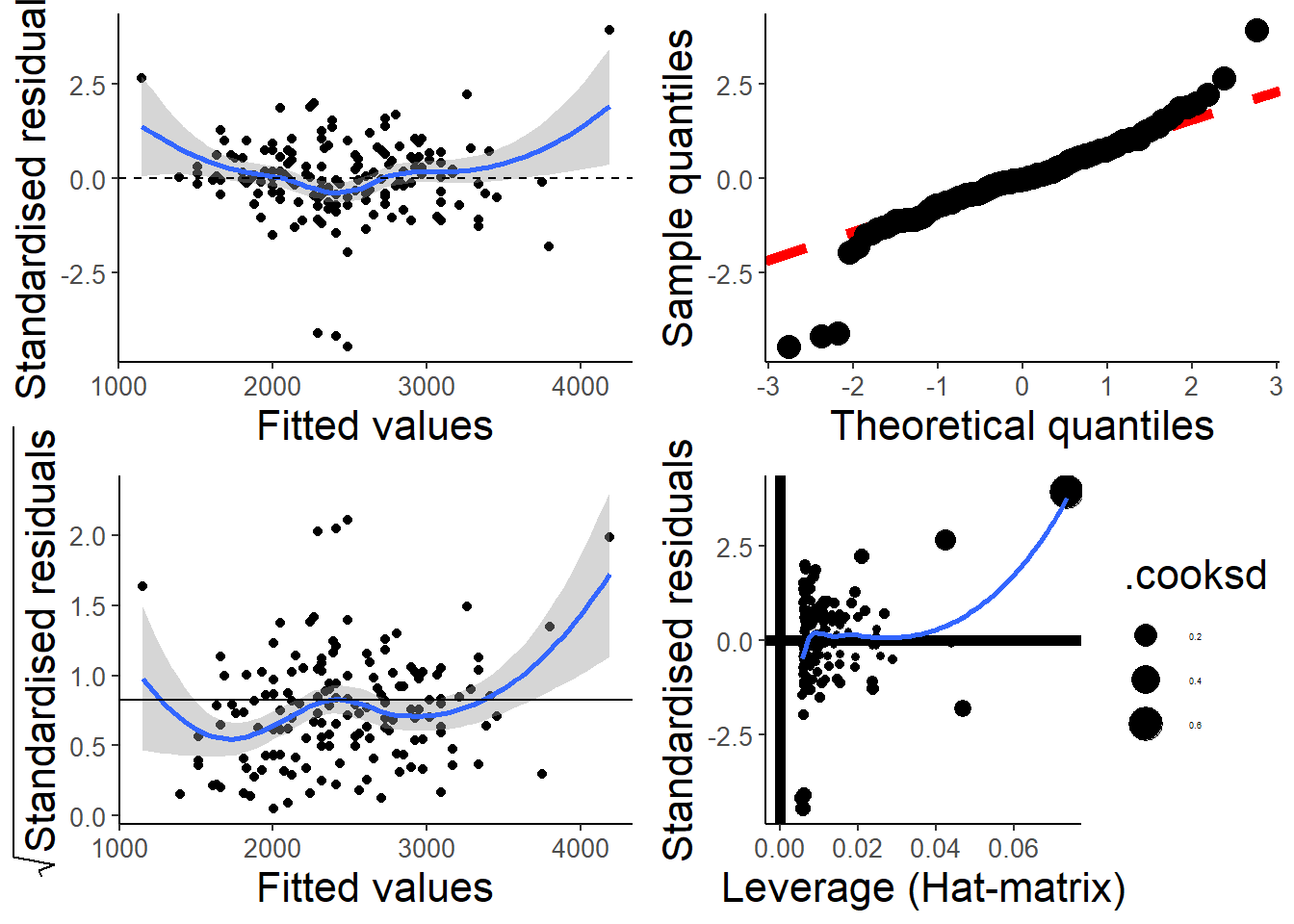
One thing that plot makes clear is the presence of several
outliers. In The Ecological Detective, Ray Hilborn and Marc Mangel
have an entire section entitled “Don’t let outliers ruin your life”.
In that spirit, the next few plots identify and eliminate the
outliers, and then refit the model. I’m going to switch
over to using broom functions here, because too easy.
I think in the video I remove the points with the largest
absolute values of the residuals, which isn’t necessarily
a good definition of an outlier.
crabs <- augment(hsc1) # add model diagnostics
#which points have biggest residuals
crabs |> ## native pipe!!
rownames_to_column()|>
arrange(desc(abs(.resid))) |>
head()## # A tibble: 6 x 9
## rowname weight width .fitted .resid .hat .sigma .cooksd .std.resid
## <chr> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 69 1300 26.5 2486. -1186. 0.00583 252. 0.0581 -4.45
## 2 26 1300 26.2 2413. -1113. 0.00579 254. 0.0508 -4.18
## 3 79 1200 25.7 2292. -1092. 0.00625 255. 0.0528 -4.10
## 4 141 5200 33.5 4184. 1016. 0.0736 256. 0.618 3.95
## 5 14 1850 21 1151. 699. 0.0425 263. 0.158 2.67
## 6 91 3850 29.7 3262. 588. 0.0209 264. 0.0526 2.22So rows 69, 26, 79, and 141 have large residuals, more than 50% larger than the remainder.
outliers <- c(26, 69, 79, 141)
hsc2 <- lm(weight~width, data = crabs[-outliers,])
plot(weight~width, data = crabs)
points(crabs[outliers,2, drop = TRUE], crabs[outliers,1, drop = TRUE], pch = "x", col = "red")
abline(hsc2, lwd = 2)
abline(hsc1, lty = 2)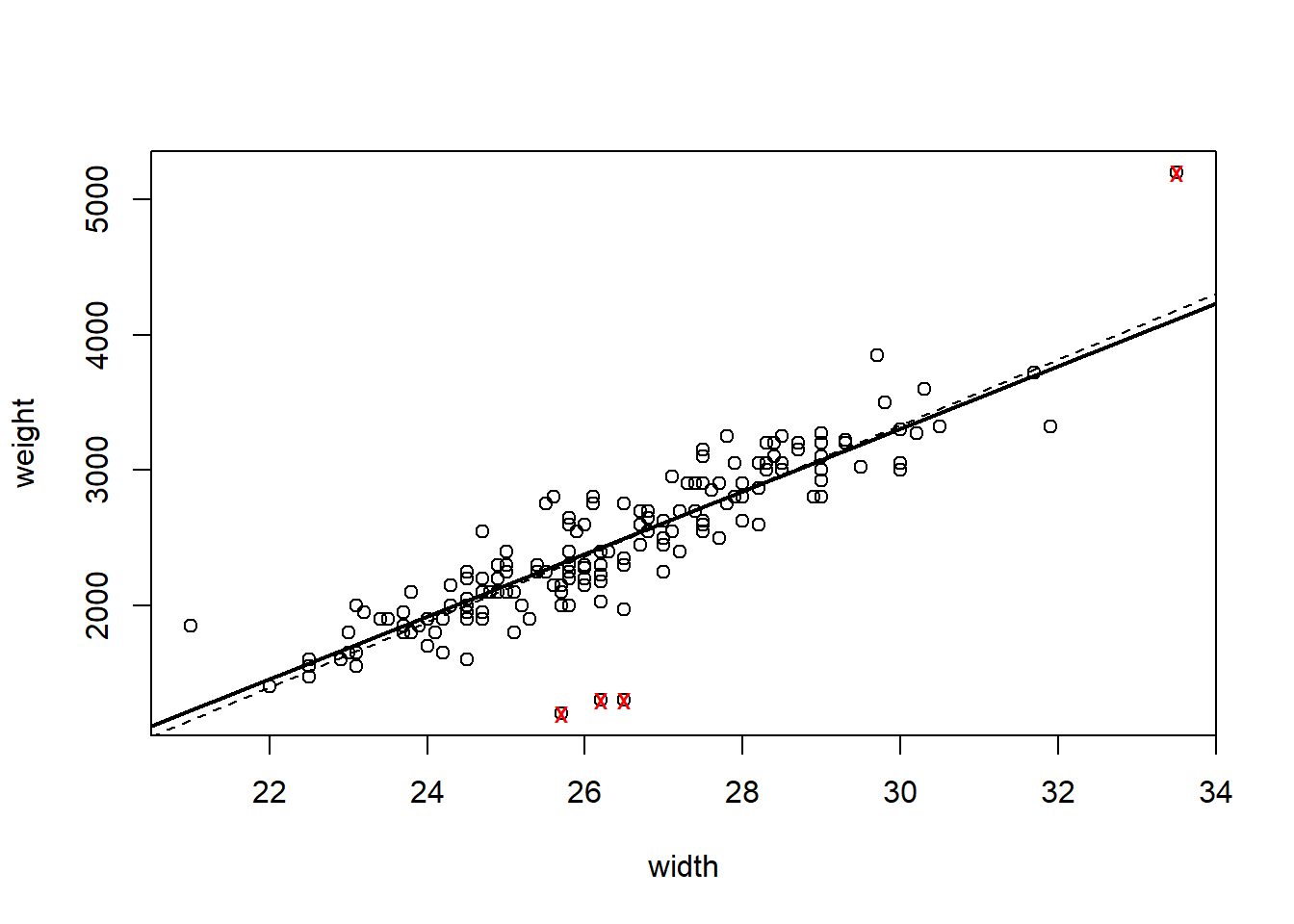
The “outliers” are identified with the red ’x’s. The original fit is given with a dashed line. How do the residual plots look with the outliers removed?
qqnorm(residuals(hsc2))
qqline(residuals(hsc2))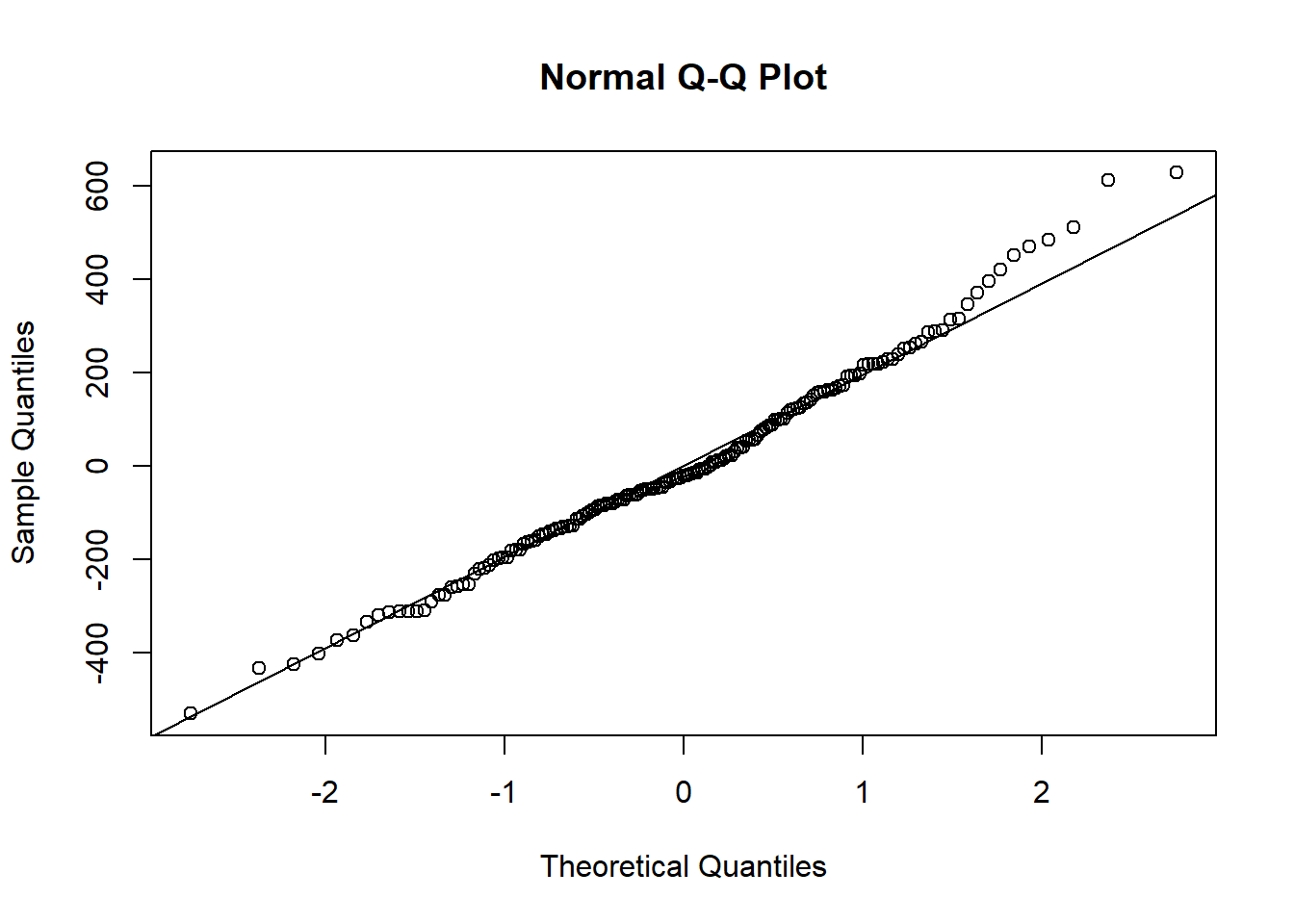
and checking for non-linearity by plotting residuals against the independent variable width.
plot(crabs[-outliers, "width", drop = TRUE], residuals(hsc2),
xlab = "width [cm]", ylab = "Residuals",
main = "Residuals vs. Independent")
abline(h = 0, lwd = 2, lty = 2)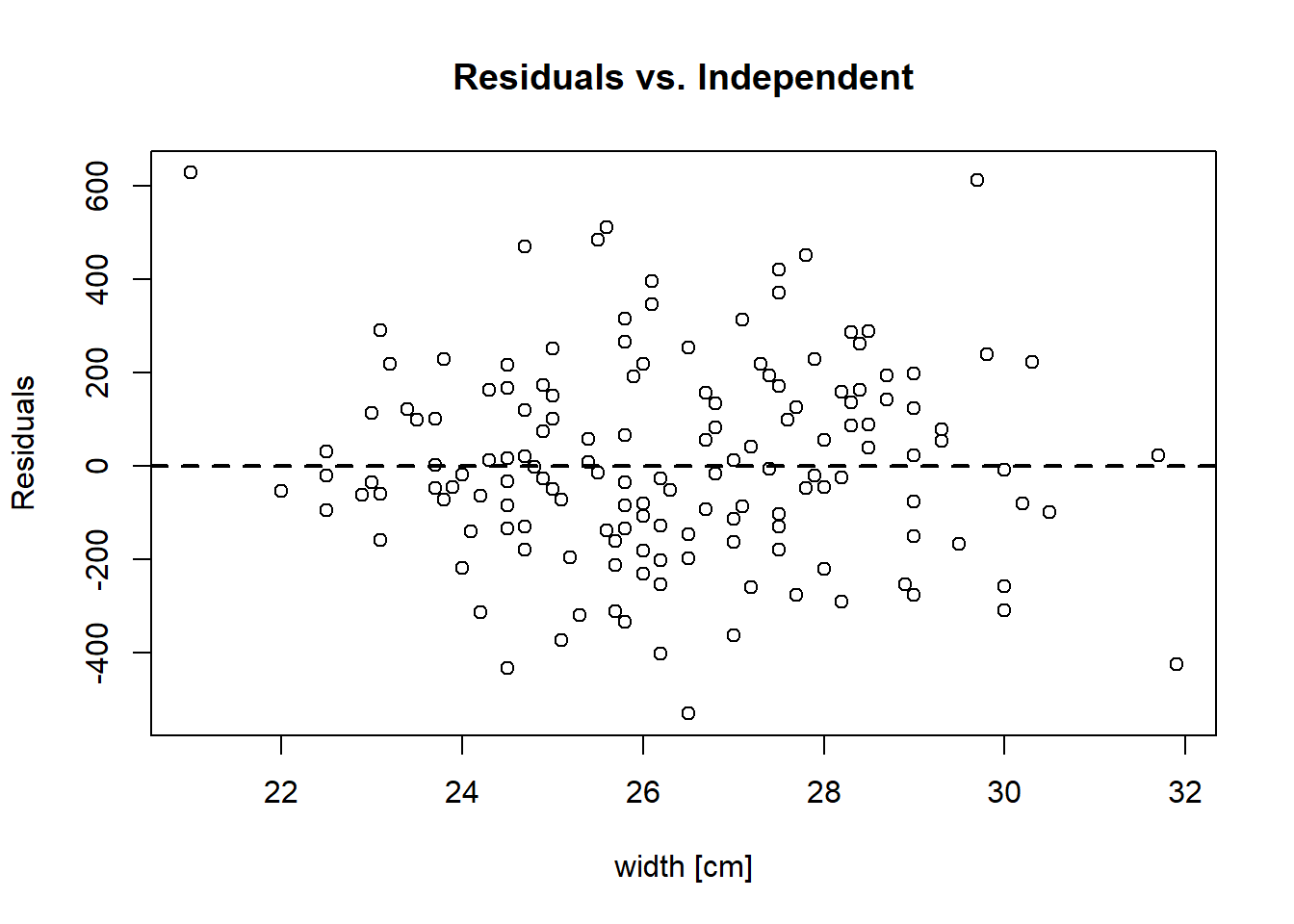
and finally with the function from my package.
check_assumptions(hsc2)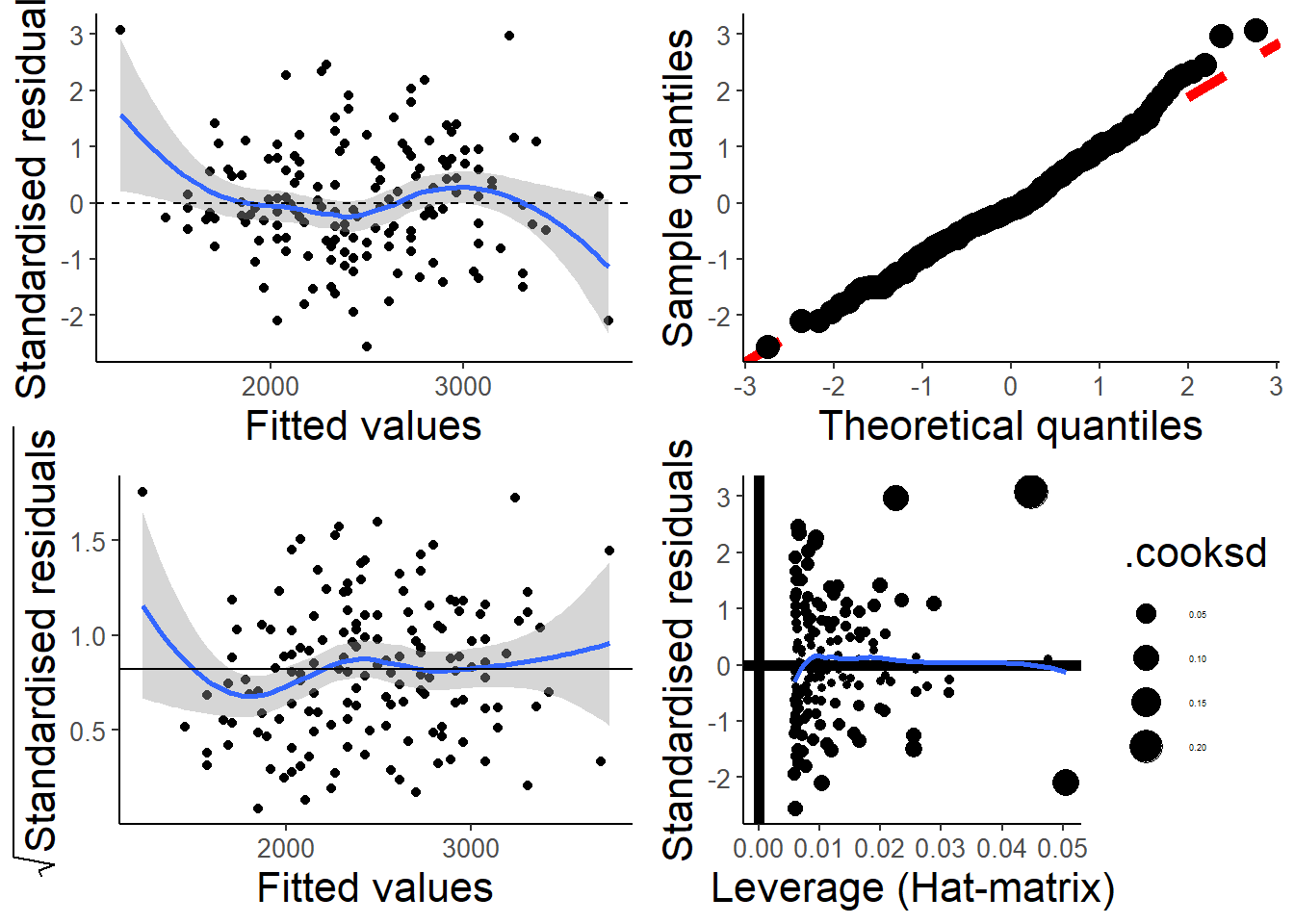
The scale-location plot in the bottom left also has a “guide” line at ~0.82, which turns out to be the expected value of the square root of the absolute value of the standardized residuals.
My students and I also developed a function that automatically plots the residuals against all the independent variables to check for possible non-linearities.
check_nonlinear(hsc2)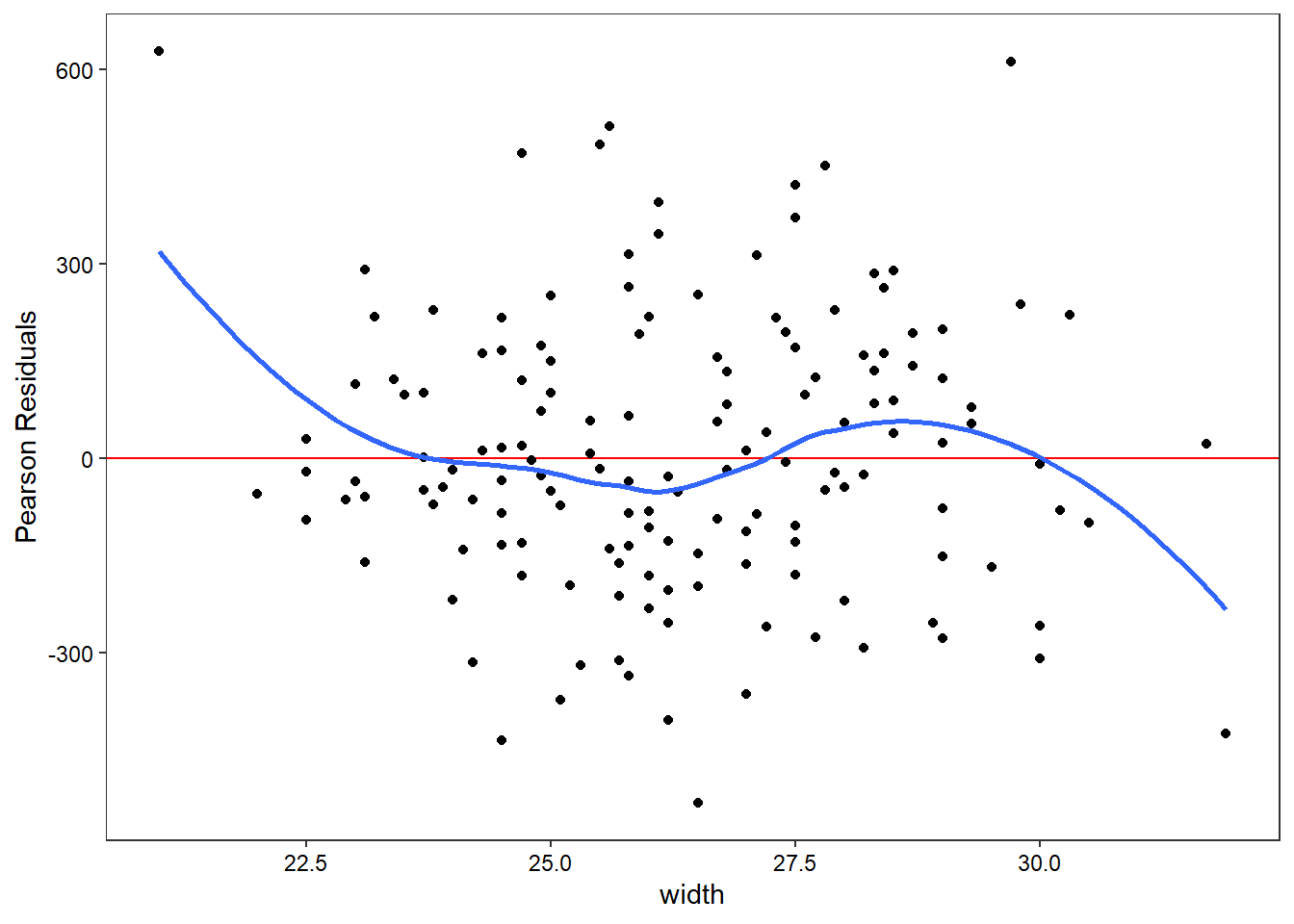
Hm. Clearly need to update that into ggplot for nice confidence regions!
